Future Research Interests
Research Overview
During my PhD, I focused on tumor diagnosis and treatment based on splicing dysregulation. My research addresses two key questions:
(1) How can we reliably identify splicing events linked to cancer progression?
(2) How can we utilize splicing dysfunction to enhance diagnostic and treatment strategies?
Through multi-omics analysis and experimental validation, I aim to translate basic research findings into clinical applications.
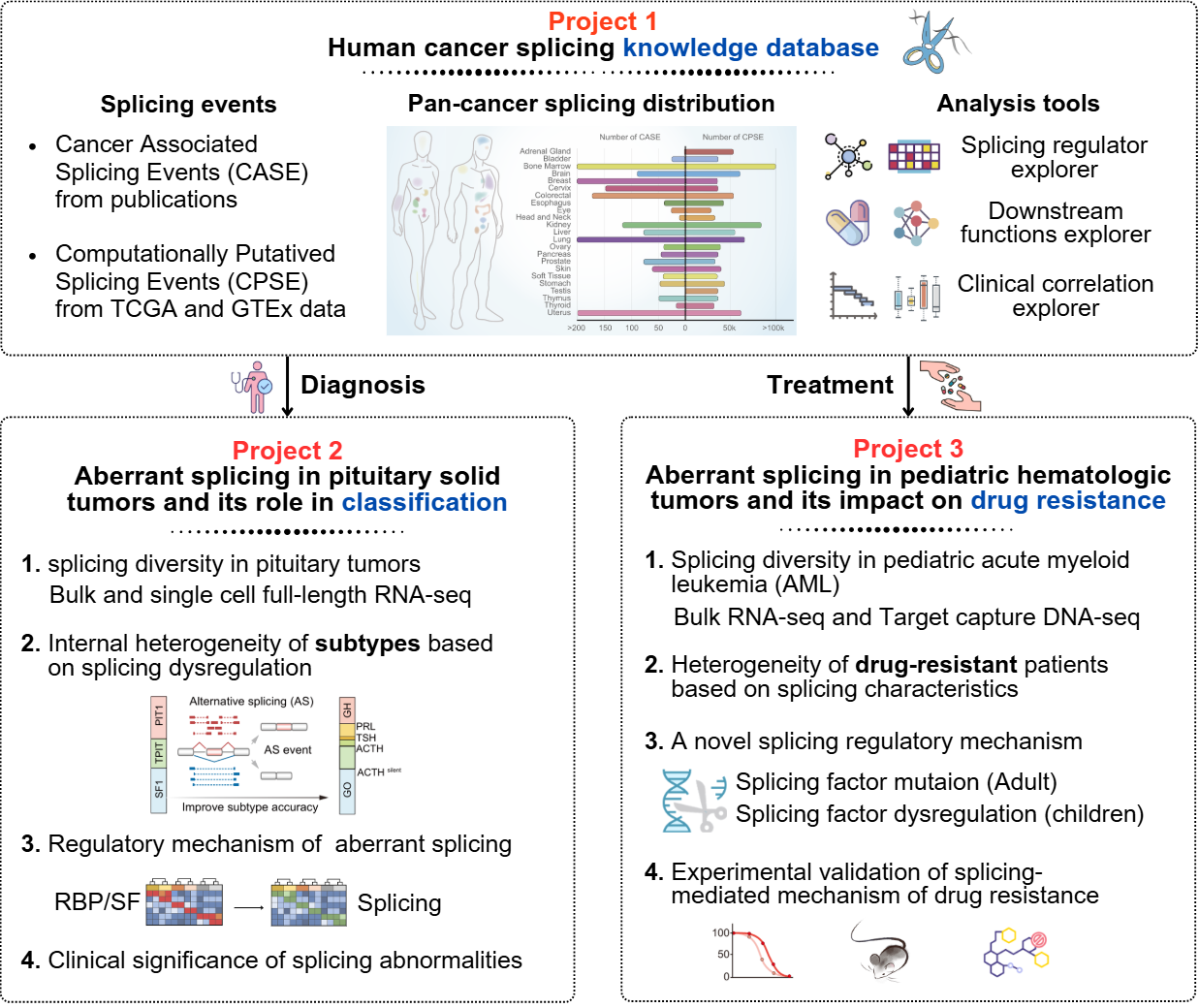
Lead Projects
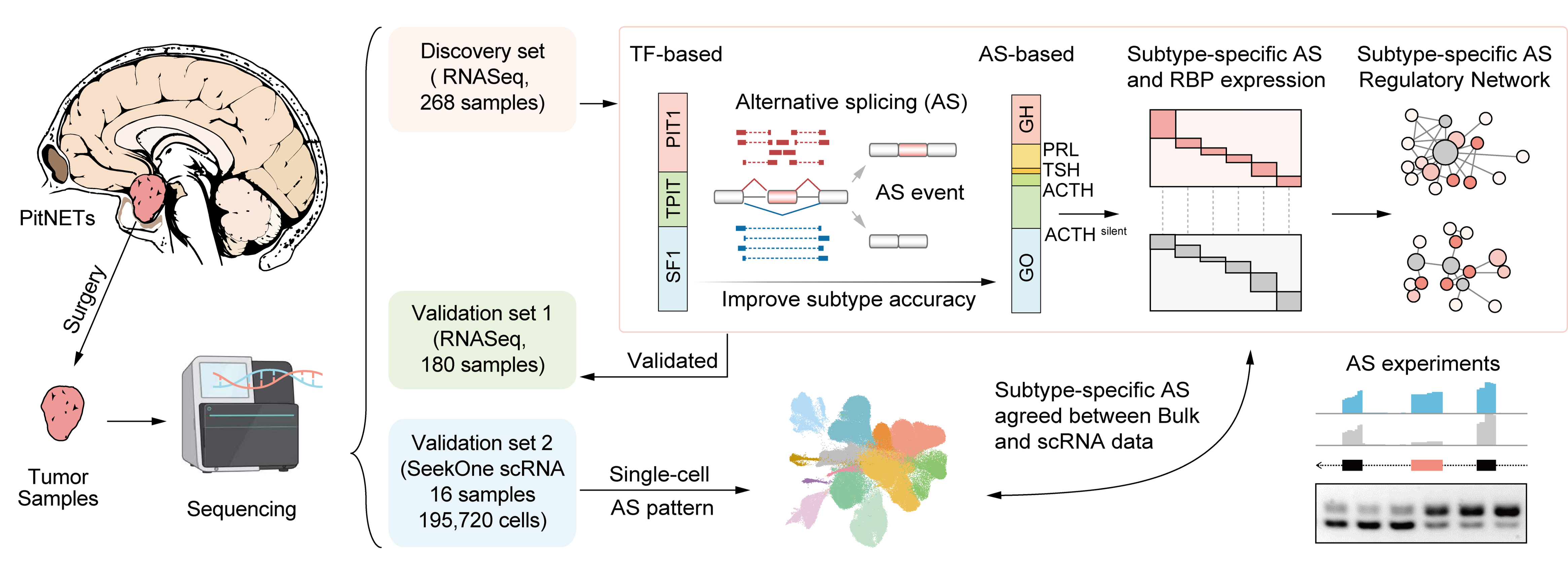
PitNETs Subtyping
PitNETs are among the most common intracranial tumors, consisting of various hormone-secreting cells with complex subtypes and clinical manifestations. Existing classifications can differentiate the three major lineages of pituitary tumors but often fail to capture their heterogeneity. This study explores alternative splicing features for precise diagnostic classification.
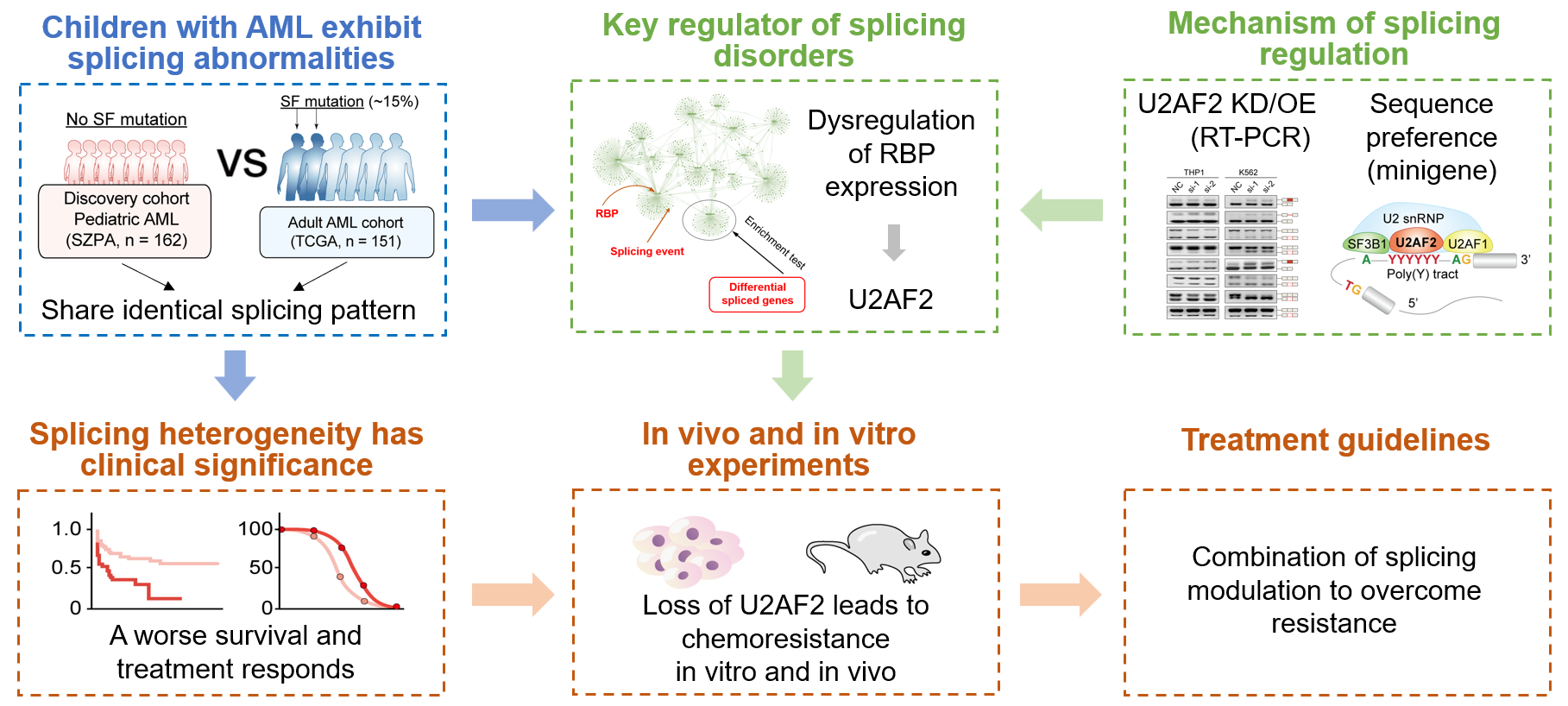
PAML Resistance
Pediatric AML is a highly aggressive malignancy, with around 30% of patients experiencing chemoresistance or relapses, often due to unknown mechanisms. While about 30% of adult AML patients have splicing factor (SF) mutations linked to splicing dysregulation, such mutations are rare in pediatric AML, leaving the role of splicing function unclear.
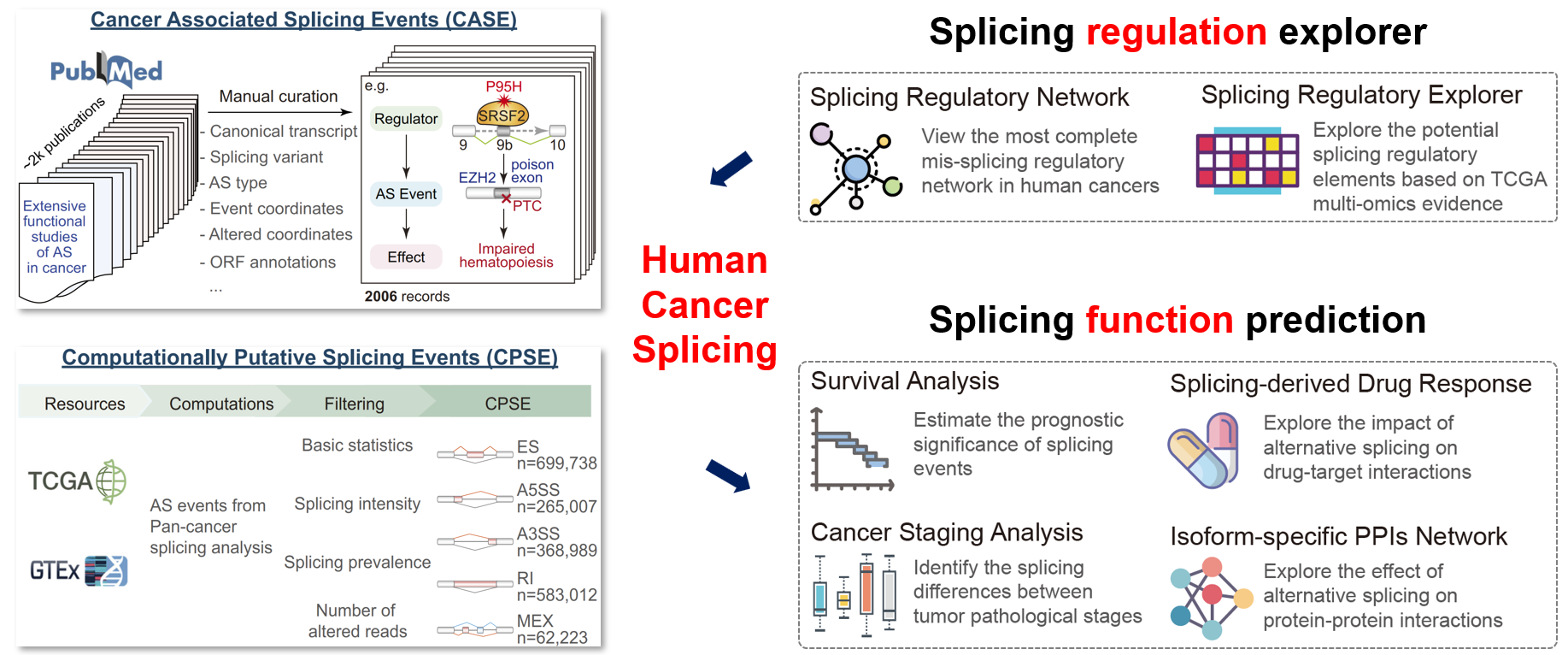
ASCancer Atlas
Systematic identification of alternative splicing events largely relies on computational methods, making it challenging to filter out true key abnormal splicing events with oncogenic potential from the vast number of computationally predicted events.
Collaborative Projects
The role of micronuclei in splicing dysregulation and cancer progression
Our study shows that the loss of splicing regulatory factors in micronuclei causes widespread splicing dysregulation and inactivation of key tumor suppressor genes.
A new RNA regulatory mechanism involving RBMX and YTHDC1 interaction in AML Progression
RBMX regulates YTHDC1 condensate properties, influencing transcription dynamics and promoting leukemia progression.
Master Regulator analysis of Alternative Splicing (MRAS)
A computational method that identifies key splicing regulators influencing splicing networks and cellular processes.